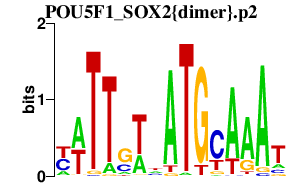
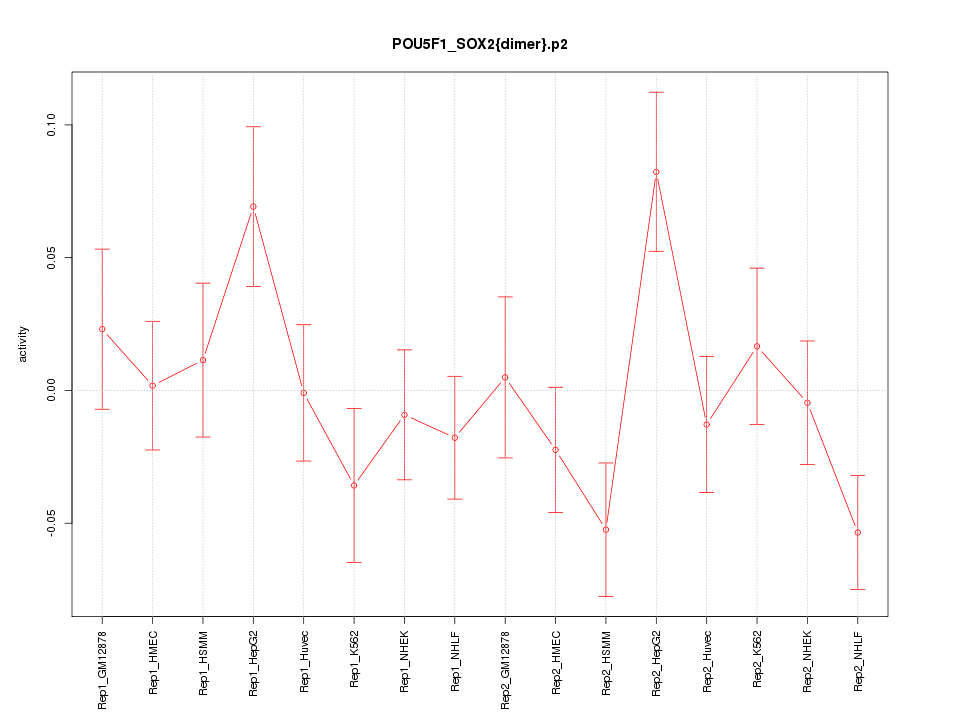
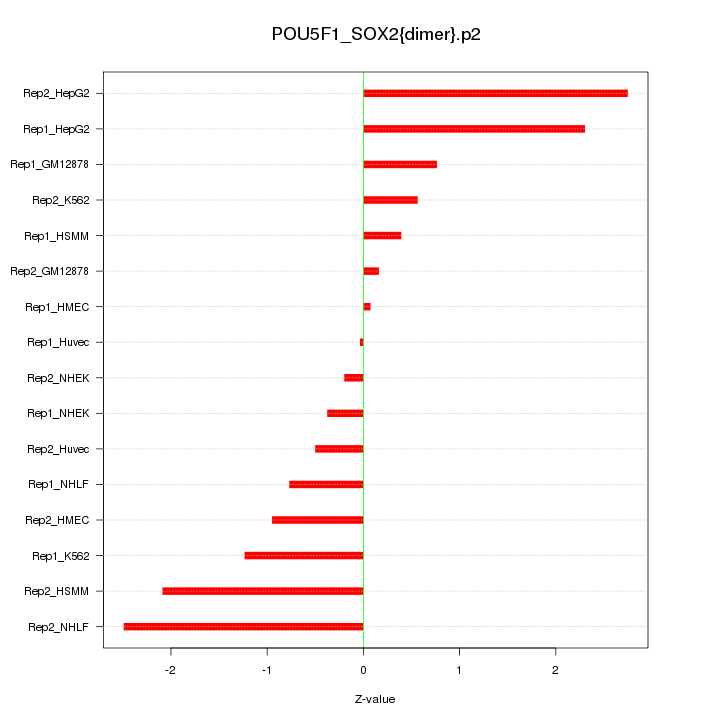
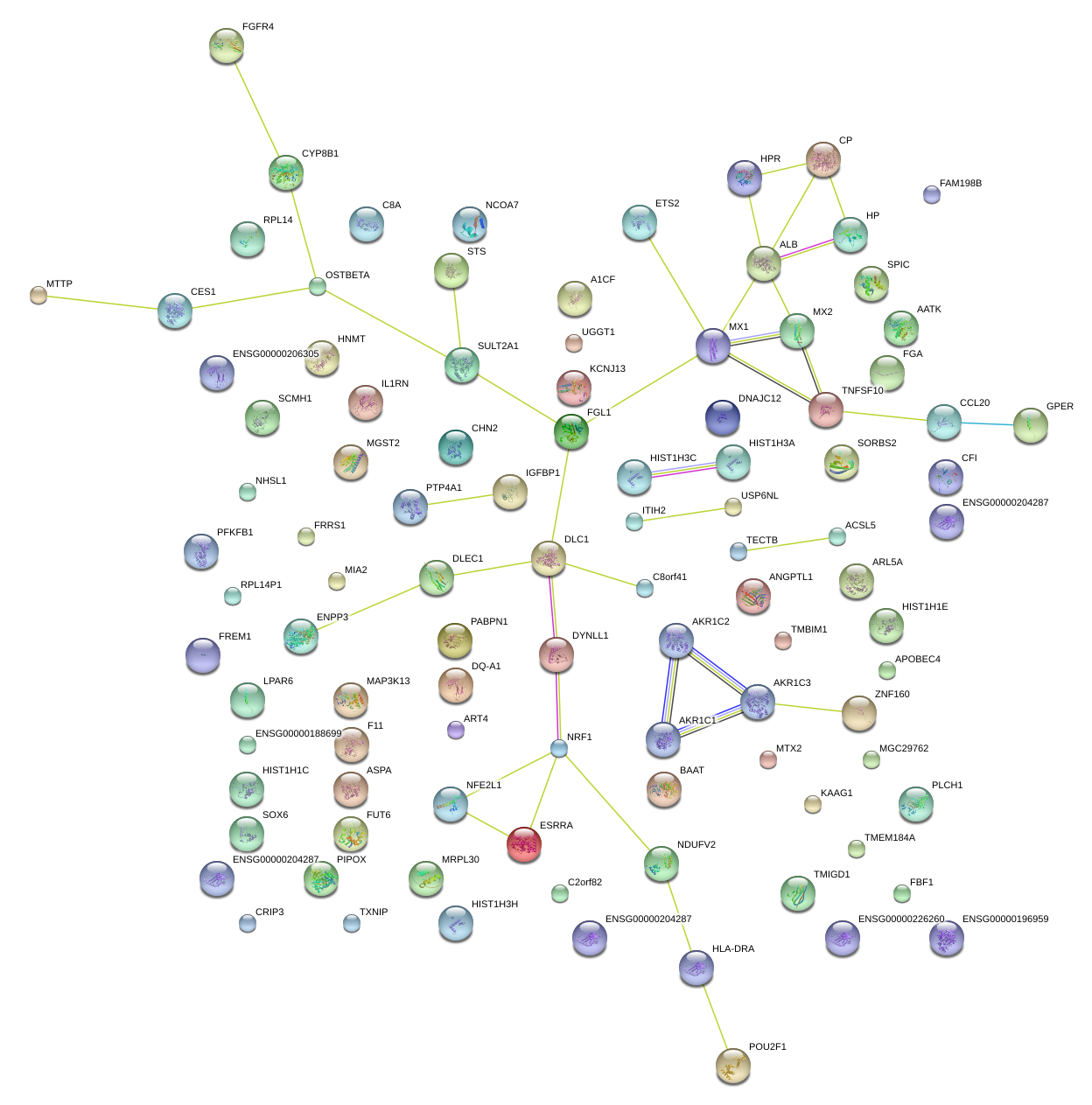
|
chr5_+_27508139
|
3.497
|
|
LOC643401
|
hypothetical protein LOC643401
|
|
chr6_-_138862273
|
2.651
|
|
|
|
|
chr10_+_7785235
|
2.437
|
NM_002216
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr17_+_24394281
|
2.424
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr8_-_17797127
|
2.401
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr4_+_100714995
|
2.400
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr10_+_7785360
|
2.370
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr10_+_7785290
|
2.327
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr10_+_7785331
|
2.315
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr17_+_24394043
|
2.232
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr2_+_128644510
|
2.210
|
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr4_+_74493918
|
2.205
|
|
ALB
|
albumin
|
|
chr21_+_41663853
|
1.933
|
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse)
|
|
chr4_+_74493174
|
1.852
|
|
ALB
|
albumin
|
|
chr3_+_186529376
|
1.824
|
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr17_+_3324096
|
1.762
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr6_+_24465109
|
1.669
|
NM_181337
|
KAAG1
|
kidney associated antigen 1
|
|
chr16_+_70646008
|
1.583
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr4_+_74493331
|
1.542
|
|
ALB
|
albumin
|
|
chr12_-_14887648
|
1.529
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chr11_-_16380967
|
1.498
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr2_+_113601616
|
1.427
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr1_+_57093030
|
1.419
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr2_+_233443237
|
1.399
|
NM_206895
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr4_+_74493266
|
1.365
|
|
ALB
|
albumin
|
|
chr13_-_47885018
|
1.355
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr19_-_5789740
|
1.334
|
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr2_+_113601608
|
1.322
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr3_-_150422474
|
1.309
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr6_+_132000134
|
1.305
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr1_+_165565006
|
1.275
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_+_74493218
|
1.262
|
|
ALB
|
albumin
|
|
chr15_+_63124760
|
1.259
|
NM_178859
|
OSTBETA
|
organic solute transporter beta
|
|
chr2_+_138438277
|
1.231
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr10_-_52315436
|
1.146
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr1_+_165564904
|
1.139
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr19_-_53081326
|
1.122
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr16_-_54424517
|
1.109
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr4_+_74493301
|
1.103
|
|
ALB
|
albumin
|
|
chr1_+_144149794
|
1.071
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr6_+_26264531
|
1.052
|
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr14_+_38772870
|
1.048
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr3_-_42892195
|
1.029
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr4_+_88076418
|
1.021
|
|
|
|
|
chr6_+_26264537
|
1.011
|
NM_005321
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chrX_-_55037235
|
0.973
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr6_+_131190237
|
0.969
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr1_-_181889070
|
0.928
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr6_-_138862271
|
0.899
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr7_+_29486010
|
0.865
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr4_-_88076328
|
0.857
|
|
|
|
|
chr4_-_110942783
|
0.846
|
NM_000204
|
CFI
|
complement factor I
|
|
chr4_+_74500748
|
0.785
|
|
ALB
|
albumin
|
|
chr10_+_114123765
|
0.785
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr1_-_177106702
|
0.781
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr14_+_22859236
|
0.777
|
NM_004643
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr4_-_186970366
|
0.773
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_+_7785347
|
0.764
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr9_-_14900992
|
0.721
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr4_+_140806536
|
0.709
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr7_+_129057152
|
0.705
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr18_+_9092618
|
0.686
|
NM_021074
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa
|
|
chr6_+_32515596
|
0.683
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr2_+_228386800
|
0.678
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chrX_+_7147471
|
0.664
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr2_-_74915722
|
0.657
|
|
|
|
|
chr10_+_4995685
|
0.644
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr7_+_45899621
|
0.643
|
|
|
|
|
chr4_+_187424336
|
0.641
|
|
F11
|
coagulation factor XI
|
|
chr6_+_64344307
|
0.619
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr6_-_43384507
|
0.608
|
NM_206922
|
CRIP3
|
cysteine-rich protein 3
|
|
chr10_-_69267774
|
0.603
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr9_-_103185621
|
0.600
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr2_-_233349469
|
0.595
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr17_-_76720342
|
0.590
|
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr11_+_63829616
|
0.582
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr8_-_33490094
|
0.575
|
NM_025115
NM_001102401
|
C8orf41
|
chromosome 8 open reading frame 41
|
|
chr2_-_218865451
|
0.572
|
NM_022152
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1
|
|
chr10_+_114123828
|
0.572
|
NM_203379
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr7_+_1092968
|
0.571
|
NM_001039966
NM_001505
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr10_-_11614279
|
0.568
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr6_+_126282078
|
0.562
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr4_-_159312973
|
0.559
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_+_140806371
|
0.551
|
NM_002413
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr4_+_187424111
|
0.547
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr2_+_176842368
|
0.543
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr5_+_176449156
|
0.537
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr2_-_152392935
|
0.526
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr1_-_100003936
|
0.521
|
NM_001013660
|
FRRS1
|
ferric-chelate reductase 1
|
|
chr10_+_114033403
|
0.501
|
NM_058222
|
TECTB
|
tectorin beta
|
|
chr14_+_87560646
|
0.500
|
|
LOC283587
|
hypothetical protein LOC283587
|
|
chr3_-_173723754
|
0.493
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chr7_+_45899080
|
0.479
|
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr12_+_61645361
|
0.479
|
|
RPL14
|
ribosomal protein L14
|
|
chr3_-_156904690
|
0.476
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr2_+_99163968
|
0.473
|
NM_145212
|
MRPL30
|
mitochondrial ribosomal protein L30
|
|
chr21_+_39117064
|
0.473
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr7_-_1562272
|
0.468
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr4_-_155731288
|
0.465
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr17_-_25685175
|
0.463
|
NM_206832
|
TMIGD1
|
transmembrane and immunoglobulin domain containing 1
|
|
chr6_+_26153590
|
0.460
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr10_-_5035967
|
0.460
|
NM_001135241
|
AKR1C1
AKR1C2
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr1_-_41398126
|
0.454
|
NM_001172222
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr10_+_4995597
|
0.445
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr12_+_102859354
|
0.434
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr19_-_5790702
|
0.432
|
NM_000150
NM_001040701
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr2_+_143351695
|
0.430
|
|
KYNU
|
kynureninase
|
|
chr9_-_74757735
|
0.425
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr11_+_20001130
|
0.416
|
NM_001111019
|
NAV2
|
neuron navigator 2
|
|
chr12_-_121767272
|
0.415
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr13_+_23042722
|
0.414
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr14_+_38720085
|
0.413
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr9_-_85773899
|
0.401
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr3_+_173043832
|
0.399
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr12_-_9123126
|
0.393
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr2_+_68445821
|
0.386
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr1_-_24112345
|
0.385
|
NM_001841
|
CNR2
|
cannabinoid receptor 2 (macrophage)
|
|
chr1_-_149214063
|
0.384
|
NM_022075
NM_181746
|
LASS2
|
LAG1 homolog, ceramide synthase 2
|
|
chr11_+_62813947
|
0.382
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr6_+_26212082
|
0.369
|
NM_003542
|
HIST1H4C
|
histone cluster 1, H4c
|
|
chr1_+_61315378
|
0.368
|
|
|
|
|
chr2_+_190248955
|
0.367
|
NM_144708
|
ANKAR
|
ankyrin and armadillo repeat containing
|
|
chr1_+_61315516
|
0.366
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chrX_+_70255128
|
0.364
|
NM_005120
|
MED12
|
mediator complex subunit 12
|
|
chr1_+_153222393
|
0.361
|
NM_001184891
NM_001184892
NM_025207
NM_201398
|
FLAD1
|
FAD1 flavin adenine dinucleotide synthetase homolog (S. cerevisiae)
|
|
chr5_-_78317444
|
0.359
|
NM_198709
|
ARSB
|
arylsulfatase B
|
|
chr10_+_4995453
|
0.356
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr4_+_74520796
|
0.339
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr9_+_130724382
|
0.337
|
NM_001100877
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr3_-_180452338
|
0.336
|
NM_171830
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr5_+_161210280
|
0.336
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr2_-_188086612
|
0.336
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr20_+_16312
|
0.335
|
NM_153325
|
DEFB125
|
defensin, beta 125
|
|
chr20_-_23534512
|
0.334
|
NM_001008693
|
CST9
|
cystatin 9 (testatin)
|
|
chr22_+_18318459
|
0.334
|
NM_001135161
|
COMT
|
catechol-O-methyltransferase
|
|
chr14_-_106241445
|
0.333
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr12_+_111829191
|
0.333
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr10_+_101532450
|
0.333
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr3_-_102878301
|
0.332
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr11_+_8016755
|
0.331
|
NM_003320
|
TUB
|
tubby homolog (mouse)
|
|
chr4_-_39199527
|
0.331
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr6_-_31668476
|
0.329
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr1_+_153222464
|
0.329
|
|
FLAD1
|
FAD1 flavin adenine dinucleotide synthetase homolog (S. cerevisiae)
|
|
chr13_+_96592051
|
0.326
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr14_+_31868229
|
0.325
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr6_-_108252200
|
0.324
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr14_-_106330781
|
0.321
|
|
IGHV5-78
|
immunoglobulin heavy variable 5-78 pseudogene
|
|
chr2_-_231697999
|
0.321
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr18_+_27425727
|
0.321
|
NM_000371
|
TTR
|
transthyretin
|
|
chr1_+_168381811
|
0.320
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr6_-_116707778
|
0.314
|
|
TSPYL1
|
TSPY-like 1
|
|
chr2_+_68445936
|
0.313
|
|
PLEK
|
pleckstrin
|
|
chr17_-_38385511
|
0.301
|
NM_025267
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chr3_-_9895633
|
0.300
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr3_+_45902999
|
0.299
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr10_-_73518772
|
0.294
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr1_-_180627934
|
0.294
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr6_+_131936057
|
0.290
|
NM_000045
|
ARG1
|
arginase, liver
|
|
chr7_+_80105907
|
0.284
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr6_+_64414389
|
0.284
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr1_+_149877955
|
0.283
|
|
SNX27
|
sorting nexin family member 27
|
|
chr11_-_55987452
|
0.281
|
NM_001004743
|
OR5M9
|
olfactory receptor, family 5, subfamily M, member 9
|
|
chr4_-_123761615
|
0.279
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr16_+_82540172
|
0.279
|
NM_013370
NM_182980
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr4_-_41445479
|
0.279
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr2_-_152393128
|
0.279
|
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr2_+_132196533
|
0.278
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr2_-_43919392
|
0.276
|
NM_022436
|
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5
|
|
chr12_+_7833261
|
0.276
|
NM_024865
|
NANOG
|
Nanog homeobox
|
|
chr17_-_15463742
|
0.274
|
NM_006382
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr3_+_109504021
|
0.267
|
NM_007072
|
HHLA2
|
HERV-H LTR-associating 2
|
|
chr12_+_111829121
|
0.267
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr2_+_99164064
|
0.266
|
|
MRPL30
|
mitochondrial ribosomal protein L30
|
|
chr16_+_52081836
|
0.264
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr2_-_180135559
|
0.262
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr9_-_13240313
|
0.258
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chr19_-_45141473
|
0.257
|
|
LOC440525
PRR13
|
proline rich 13 pseudogene
proline rich 13
|
|
chr7_+_80105718
|
0.254
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chrX_+_128741578
|
0.253
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr9_+_130485926
|
0.251
|
|
SET
|
SET nuclear oncogene
|
|
chr14_-_94053933
|
0.247
|
NM_173850
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12
|
|
chr5_+_68702617
|
0.247
|
NM_133340
NM_133341
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr14_-_34414603
|
0.245
|
NM_013448
NM_182648
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr1_-_93172119
|
0.245
|
|
|
|
|
chr10_+_91488704
|
0.244
|
|
KIF20B
|
kinesin family member 20B
|
|
chr9_+_2707525
|
0.240
|
NM_133497
|
KCNV2
|
potassium channel, subfamily V, member 2
|
|
chr16_+_28850760
|
0.239
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr10_+_124141808
|
0.239
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr12_+_25096472
|
0.239
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr15_-_32398220
|
0.237
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr20_-_18425811
|
0.237
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr2_+_143351536
|
0.237
|
NM_001032998
NM_003937
|
KYNU
|
kynureninase
|
|
chr4_+_185063421
|
0.236
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr19_+_59820413
|
0.236
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr6_-_27890496
|
0.235
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr12_+_9713472
|
0.231
|
NM_001197318
NM_013269
NM_001004419
NM_001197317
NM_001197319
|
CLEC2D
|
C-type lectin domain family 2, member D
|
|
chr7_-_122126422
|
0.230
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr11_+_20000938
|
0.230
|
|
NAV2
|
neuron navigator 2
|
|
chr6_+_167445284
|
0.229
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr2_-_37397669
|
0.229
|
NM_005813
|
PRKD3
|
protein kinase D3
|